Apheris Registry: nnU-Net🔗
nnU-Net is an open-source model for biomedical image segmentation, based on the 2020 paper by Isensee et. al. It is designed to configure itself for various datasets without manual intervention. By optimizing its entire pipeline for each new dataset, including preprocessing, network architecture, training, and post-processing, nnU-Net aims to address the complexities of creating deep learning models for medical image analysis.
Federating nnU-Net on Apheris🔗
To bring nnU-Net into the Apheris environment, we use the pip-installable package for nnU-Net and add wrapping code to federate it use NVIDIA FLARE.
Fingerprinting, Planning and Pre-Processing🔗
nnU-Net differs from many machine learning models in that it takes an Auto-ML approach to determine the model structure and any pre-processing required on the input data.
To do this, it uses a technique called fingerprinting; essentially calculating a set of statistics over the dataset, which serves as input to the planning phase, where it makes decisions on various aspects of the model structure. For more details, please see the nnU-Net paper.
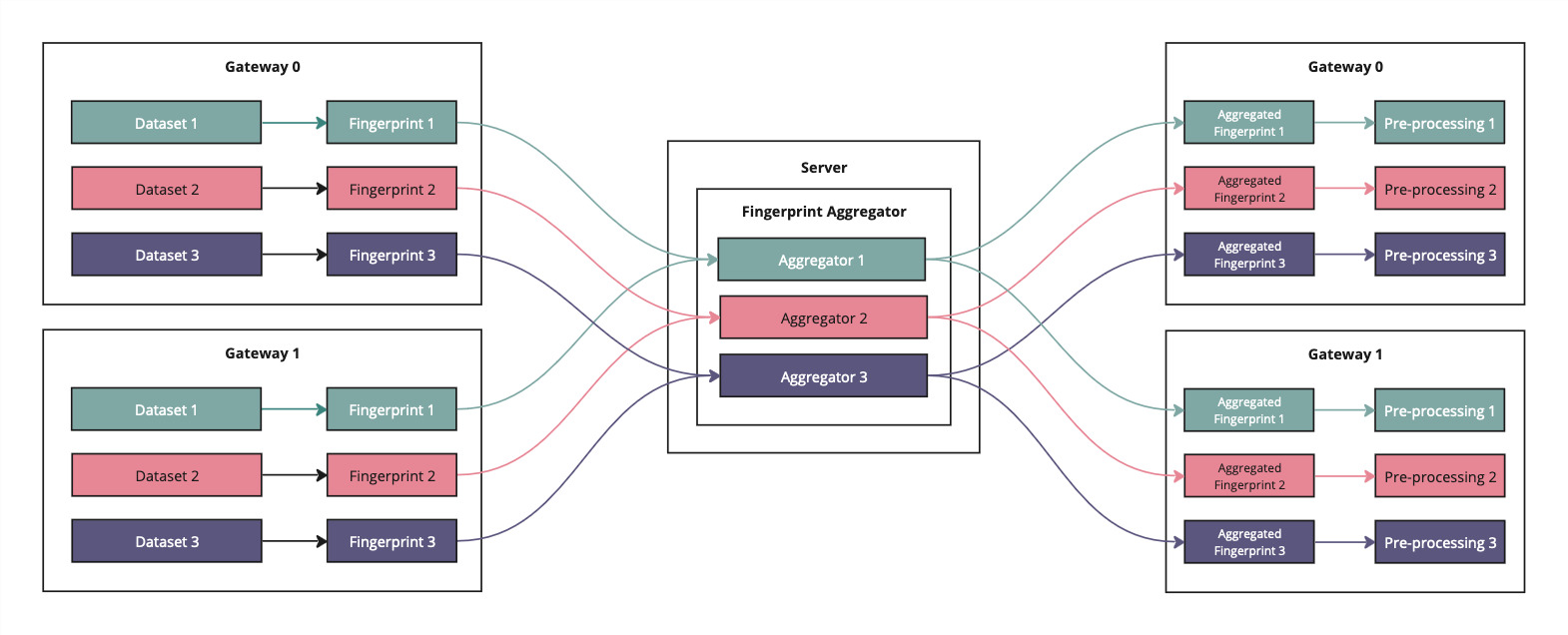
To federate nnU-Net, Apheris nnU-Net must federate these fingerprint statistics, such that the resulting model takes into account the statistics of the overall combined dataset from all sites. The statistics themselves are sufficiently abstracted from the raw data, that there are no privacy concerns with sharing them with the Orchestrator.
Therefore, we carry out fingerprinting on each Apheris Gateway individually, then pass the fingerprints back to the Orchestrator for aggregation. The fingerprint statistics are then aggregated on the Orchestrator and returned to the client site where they are combined with the dataset descriptors that nnU-Net generates.
Once they receive the aggregated fingerprints from the orchestrator, the Gateways will start the planning phase. This is deterministic with respect to the fingerprint input, therefore we expect all gateways to produce the same plan. There is a sanity check within the code, that will ensure this is the case, since it is important that all Gateways operate on the same model.
Finally, once we are confident that all plans are identical, we perform the preprocessing defined by the plan against the datasets in each Gateway. The pre-processed data remains on the Gateway for the duration of the Gateway's activation.
Since this functionality is slightly beyond the standard training loop for machine learning, Apheris nnU-Net Uses a number of custom FLARE components to enable this.
Training🔗
Once the data is pre-processed, it is simple to train the model using nnU-Net's training functions. After one or more epochs; we load the weights, return them to the server, aggregate them and pass the aggregated model back to each Gateway. nnU-Net is a parametric model, and therefore we can apply FedAvg Federation using the built-in NVIDIA FLARE ScatterAndGather component.
Once the requested number of rounds have completed, you can download the resulting checkpoint from the Orchestrator using the CLI. The checkpoint will be formatted for nnU-Net, with the appropriate metadata for inference.
Inference🔗
Apheris nnU-Net also supports federated inference. In this case, the model weights are kept on each Gateway alongside the data. For each dataset, we run prediction, compile the results into a zip archive, and return them to the Orchestrator. The user can then download them with the NVIDIA FLARE workspace upon completion of their run. The checkpoint must be of a compatible format for nnU-Net, and must already reside in the Gateway's Docker image. For more information on how to embed you checkpoint into your Gateway image, please contact an Apheris representative.
Validation🔗
The model also supports federated evaluation, during which the validation accuracy is calculated at each Gateway on a hold-out evaluation set. Then, it is aggregated in the Orchestrator and stored in the workspace to be returned to the user.
The checkpoint must be of a compatible format for nnU-Net, and must already reside in the Gateway's Docker image. For more information on how to embed you checkpoint into your Gateway image, please contact an Apheris representative.
Datasets🔗
When training nnU-Net, your data needs to be organised in a specific format to allow it to be ingested correctly into the model. We assume that this data pre-formatting step has been undertaken before the dataset has been registered to Apheris. For more information on the specific format, please see the nnU-Net documentation.
Important
Please note that a federated computation across multiple datasets requires that each dataset resides in a different Gateway.
Parameters for Apheris nnU-Net🔗
To run Apheris nnU-Net, you need to provide a set of parameters that tell Apheris how to execute your model. In this section, we'll show you what those parameters are, and how you can use them to customise your federated workloads on Apheris.
When using the Apheris CLI, you provide parameters as a JSON payload. This payload should match the schema that is used in the model definition, to ensure your workload runs without error.
Training Payload🔗
The payload is defined by this Pydantic model (no other fields are allowed):
class TrainingPayload(BaseModel):
mode: Literal["training"]
num_rounds: int
dataset_id: int
model_configuration: Literal["2d", "3d_lowres", "3d_fullres"]
device: Literal["cpu", "cuda", "mps"] = "cuda"
fold_id: Union[Literal["all"], int] = "all"
trainer_name: str = "nnUNetTrainer"
epochs_per_round: int = 1
starting_fed_round: int = 0
num_rounds is the number of communication rounds for your training run.
dataset_id is the nnU-Net dataset ID, i.e. XYZ in DatasetXYZ_name_text.
For example, if the dataset you wish to use is Dataset004_SiteA_Data, the dataset ID
is 4.
model_configuration is the nnU-Net model configuration to train. Cascades are not
currently supported.
device is the hardware device on which to train. Generally this should be cuda. On
M* Apple hardware, you can use mps to accelerate training.
fold_id is the fold of the dataset to use. nnU-Net supports performing k-fold cross-validation during training. You can supply the fold number (0-4) to train only a specific
fold of the dataset (see the nnU-Net documentation
for more details). We recommend training with "all" when using federated training on
Apheris as it is not currently possible to pass the splits file between submitted jobs.
trainer_name must be a valid trainer name from the nnU-Net code-base.
epochs_per_round is the number of times we pass through the dataset between federated
aggregations.
starting_fed_round can be used if you wish to resume from an existing training run.
Example:
{
"mode": "training",
"device": "mps",
"num_rounds": "2",
"model_configuration": "2d",
"dataset_id": 4
}
Inference Payload🔗
The payload is defined by this Pydantic model (no other fields are allowed):
class InferencePayload(BaseModel):
mode: Literal["inference"]
checkpoint_path: Path
fold_id: Union[Literal["all"], int]
device: Optional[Literal["cpu", "cuda", "mps"]] = "cuda"
apheris_dataset_subdirs: Dict[str, str] = {}
checkpoint_name: Literal["checkpoint_final.pth", "checkpoint_best.pth"] = (
"checkpoint_final.pth"
)
checkpoint_path is the location of the checkpoint on the Gateway. Note that it is
currently necessary to build a custom model image containing the checkpoint.
fold_id is the fold of the dataset to use - the dataset will be split and only the
specified part of the split will be used. This must match whatever is in your checkpoint.
device is the hardware device on which to execute. Generally this should be cuda. On
M* Apple hardware, you can use mps to accelerate inference.
apheris_dataset_subdirs for datasets provided as IDs (as they will be in remote data
runs), you can provide a subdirectory path inside the dataset that contains the data.
For example, if your dataset is an nnU-Net formatted dataset, the inference data is inside
imagesTs. This is a dictionary of Apheris Dataset ID to subdirectory. The subdirectory
path must not be absolute and cannot be outside the dataset root or the run will raise
an error.
checkpoint_name whether to use the best or final weights from training. This must
match whatever is in your checkpoint.
Example:
{
"mode": "inference",
"device": "cpu",
"fold_id": 0,
"checkpoint_path": "/local/workspace/nnUNet_results/Dataset001_XYZ/nnUNetTrainer__nnUNetPlans__2d",
"apheris_dataset_subdirs": {
"medical-decathlon-task004-hippocampus-a_gateway-1_org-1": "imagesTs"
}
}
Validation Payload🔗
It is possible to run to evaluate a checkpoint on a dataset.
This will use the checkpoint provided in the payload and run validation over the
dataset (datasets if using multiple Gateways) with nnU-Net dataset ID equal to dataset_id.
The results will be aggregated in the Orchestrator and saved to the server workspace in a file:
<server_workspace>/app_server/validation_results.json
Currently, validation only measures the DICE score as that is the only validation metric output to the logs.
Standalone validation on a pretrained checkpoint can be run using this Pydantic model:
class ValidationPayload(BaseModel):
mode: Literal["validation"] = "validation"
checkpoint_path: Path
dataset_id: int
model_configuration: Literal["2d", "3d_lowres", "3d_fullres"]
device: Literal["cpu", "cuda", "mps"] = "cuda"
fold_id: Union[Literal["all"], int] = "all"
trainer_name: str = "nnUNetTrainer"
checkpoint_path is the location of the checkpoint inside the Apheris nnU-Net container.
The checkpoint should be built into the nnU-Net image using the
Apheris Custom Model workflow.
If running using the simulator, this should be the path to the checkpoint on your local
file system.
dataset_id is the nnU-Net dataset ID, i.e. XYZ in DatasetXYZ_name_text.
For example, if the dataset you wish to use is Dataset004_SiteA_Data, the dataset ID
is 4.
model_configuration is the nnU-Net model configuration to train. Cascades are not
currently supported.
device is the hardware device on which to train. Generally this should be cuda. On
M* Apple hardware, you can use mps to accelerate training.
fold_id is the fold of the dataset to use, which should match the fold you wish to use
from the checkpoint.
trainer_name must be a valid trainer name from the nnU-Net code-base.
Resources:
- You can find the GitHub here: https://github.com/MIC-DKFZ/nnUNet
- The paper is available here: https://www.nature.com/articles/s41592-020-01008-z
Next steps🔗
Now you know a bit more about Apheris nnU-Net, why not try it out by following our guide on Machine Learning with Apheris using the NVIDIA FLARE Simulator and Python CLI?