ApherisFold Application🔗
Welcome to the ApherisFold Application. This guide will walk you through the steps needed to deploy, configure, and begin using the application in your local or cloud environment.
Overview🔗
The ApherisFold Application allows you to predict the 3D structures of macromolecule complexes with an emphasis on protein–ligand, protein–protein, and antibody–antigen interfaces using OpenFold3 (OF3) and Boltz-2. The affinity prediction capabilities of Boltz-2 are retained.
Key capabilities include:
- Quick deployment to your local environment, private cloud, or on-premises infrastructure using Docker or Kubernetes
- Model prediction (for both OF3 and Boltz-2) with input validation, batch execution programmatically or via a scientist-friendly GUI with built-in visualizations (pLDDT, PAE, structure viewers)
- Benchmarking to evaluate model performance against ground-truth structures with quantitative metrics
We will soon release additional support for data preparation and fine-tuning.
System Overview🔗
Apheris has created Docker images for its models, built on open-source versions and equipped with an HTTP API wrapper for easy integration with third-party systems.
Models and Requirements🔗
OpenFold3 and Boltz-2 are included in the initial release of the ApherisFold application. Both models are large and can require 25GB of disk space or more. These models have been packaged by Apheris as Docker images and can be freely downloaded. The images can be pulled in advance if pulling from our repository is not an option for your environment. Please contact us for assistance with this process if needed.
We target the latest release from the respective model providers. To see which models and versions your deployment is currently using, open the Models page in the Hub (see Models).
Recommended Hardware:
- Modern GPU with at least 48GB GPU memory and CUDA 11+ support (e.g. NVIDIA A100, H100, L40S, RTX 6000 and others). In AWS, the G6e instance is an example of a cost-effective machine that supports OpenFold3.
- At least 300GB of disk space
- Docker environment with Nvidia GPU drivers and the Nvidia Container Toolkit installed
Architecture🔗
The Apheris Hub operates as a coordinator over one or more model services that you deploy alongside it (using the Docker deployment script or Kubernetes Helm chart). Each model runs as its own long-lived HTTP service (for example, a Docker container or Kubernetes Pod) and exposes a standardized prediction API.
The Hub discovers model endpoints from deployment-time configuration (for example, models settings in config.yaml or models.instances in the Helm chart) and routes prediction requests to those endpoints. GPU and CPU resources are managed by your container runtime or cluster scheduler.
In Docker deployments, substantial GPU resources are only allocated while a prediction job is actually running. When a query is submitted, the model service performs the computation and acquires the required GPU resources; once the job has completed, those resources are released so the GPU can be reused for subsequent jobs.
In Kubernetes deployments using the Helm chart, each GPU-backed model runs as a long-lived Pod that requests a full GPU. That Pod retains exclusive access to the requested GPU while it is running, even when no prediction jobs are active. To run multiple GPU-backed models concurrently, use a node with more than one GPU, or deploy only one GPU-backed model at a time per single-GPU node.
You can submit multiple prediction queries at the same time. However, because each query requires full access to the GPU, they are processed sequentially.
When a query is submitted, the input is provided in JSON format as specified in the Running prediction section below, and including any assets uploaded via the "assets" option. These are saved to the input folder specified during the deployment process (see Docker Deployment or Kubernetes Deployment (Helm Chart)).
Once prediction is complete, the model's predictions and any associated logs are written to the respective output folder and immediately viewable in the UI or accessible by your own external tooling.
Getting Started🔗
Deploy the ApherisFold Application🔗
Deploying the ApherisFold Application sets up the Apheris Hub along with enabled models, and configures storage for your data and results. The ApherisFold Application can deploy using either Docker or Kubernetes.
Choose your deployment method:
- Docker Deployment - Deploy using Docker with automated deployment script
- Kubernetes Deployment (Helm Chart) - Deploy to Kubernetes using Helm
After installation, you can access the Apheris Hub from your web browser.
For step-by-step instructions, see the Troubleshooting Guide, which covers common operational problems and provides troubleshooting techniques to help identify, address, or confirm issues.
If you need help or encounter any issues, and the Hub is running, you can generate a Support ZIP archive directly from your web browser (Settings > Support > Download Support ZIP) or via the API. This archive includes system information, Hub configuration, and installed model versions that can help our team better understand your setup. Please email the Support ZIP file, along with a detailed description of your issue, to support@apheris.com.
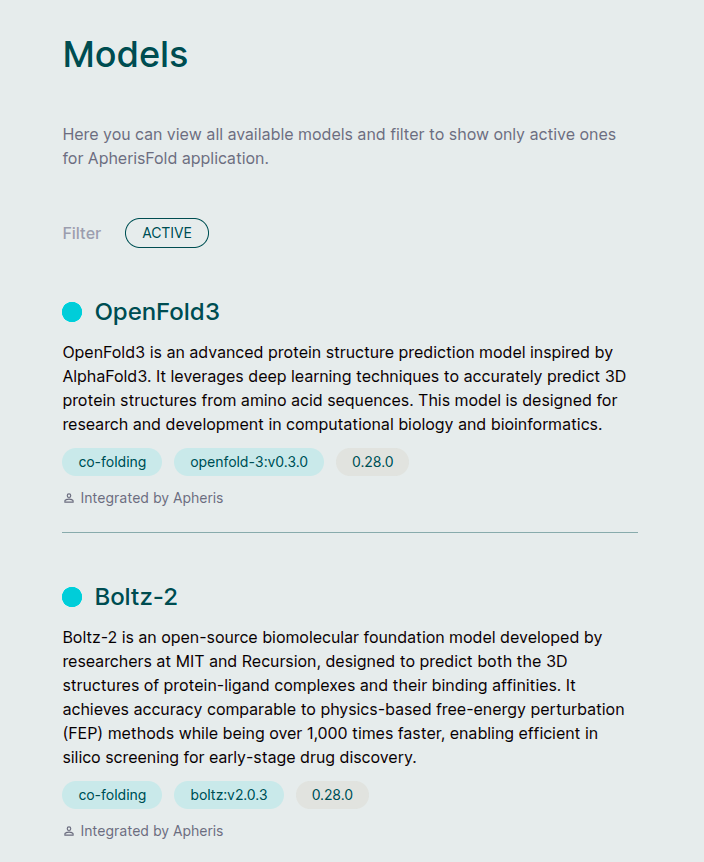
Models🔗
The Apheris Hub works with multiple models (currently OpenFold3 and Boltz-2) for the ApherisFold application. These models have been packaged by Apheris as Docker images and can be deployed in your environment. They come with a standardized query payload and can be called via an HTTP API. See Running Prediction for more information.
Which models appear in the Hub depends on how your deployment has been configured (for example, via Docker or Kubernetes). Once the Hub is running and can reach the configured model endpoints, the Models screen lists all available models and their discovered versions. Selecting a model opens its details, including descriptions and version metadata.
Mock Model🔗
When enabled, the Mock Model is useful for quickly exercising the full end-to-end ApherisFold workflow.
The Mock Model:
- Is designed to work with the built-in ApherisFold examples (and would fail on unseen inputs)
- Produces example results that are qualitatively similar to OpenFold3 outputs
- Results can be visualized and downloaded the same as any other model
- Does not require GPU resources
To enable the Mock Model, see the Docker Deployment or Kubernetes Deployment (Helm Chart) instructions.
Customizing Model Weights🔗
You can mount custom model weights into your deployment to use custom-trained or fine-tuned models. This is supported for OpenFold3 and Boltz-2.
Required Files🔗
Each model type expects specific files in the mounted weights directory:
OpenFold3:
of3_ft3_v1.pt(checkpoint file)
Boltz-2:
boltz2_conf.ckpt(confidence checkpoint)boltz2_aff.ckpt(affinity checkpoint)ccd.pkl(coordinate data)mols/(directory containing molecule definitions)
Directory Structure Examples🔗
Your weight files must be organized as follows:
OpenFold3:
weights_mount/
├── additional_weights.json
├── fed/
│ └── of3_ft3_v1.pt
└── fine-tuned/
└── of3_ft3_v1.pt
Boltz-2:
weights_mount/
├── additional_weights.json
├── custom/
│ ├── boltz2_conf.ckpt
│ ├── boltz2_aff.ckpt
│ ├── ccd.pkl
│ └── mols/
│ ├── ATP.cif
│ ├── GTP.cif
│ └── ...
└── domain-specific/
├── boltz2_conf.ckpt
├── boltz2_aff.ckpt
├── ccd.pkl
└── mols/
├── ATP.cif
├── GTP.cif
└── ...
Configuration Methods🔗
There are two ways to configure custom weights for your models:
Choose One Method
These methods are mutually exclusive. Choose either the configuration file method or the environment variable method, not both. If both are configured, the environment variable (APH_AVAILABLE_WEIGHTS) takes precedence.
Method 1: Using a Configuration File🔗
This method uses a JSON configuration file that you mount into the container. The file path is specified using the APH_WEIGHTS_CONFIG_FILE environment variable.
When to use:
- You have multiple weight sets with complex configurations
- You want to manage weights configuration separately from deployment configuration
- You need to update weights metadata without redeploying
How it works:
- Create a JSON file with your weights configuration
- Mount the file into the container (via ConfigMap in Kubernetes, or volume mount in Docker)
- Set the
APH_WEIGHTS_CONFIG_FILEenvironment variable to point to the file path inside the container
Method 2: Using an Environment Variable🔗
This method provides the weights configuration directly as a JSON string in the APH_AVAILABLE_WEIGHTS environment variable.
When to use:
- You have a simple configuration with one or two weight sets
- You want all configuration in one place (e.g., Helm values)
- You prefer not to manage separate configuration files
How it works:
- Format your weights configuration as a JSON array
- Set the
APH_AVAILABLE_WEIGHTSenvironment variable with the JSON string - The model reads the configuration directly from the environment
JSON Configuration Format🔗
Regardless of which method you choose, the JSON structure is the same. The configuration should be a JSON object with an available_weights array, where each weight entry contains:
model_type(required): Model identifier -"openfold3"or"boltz2"version(required): Version identifier in semver format (e.g.,"3.0.0-federated")mounted_path(required): Absolute path inside the container where weight files are locateddescription(optional): Human-readable description of the weightsmodel_version_id(optional): Hub model version identifier for compatibility checks - defaults to the current image's version idmodel_version_tag(optional): Hub model version tag for compatibility checks - defaults to the current image's version tag
Please see the example below for OpenFold3 custom weights:
{
"available_weights": [
{
"model_type": "openfold3",
"version": "3.0.0-federated",
"description": "OpenFold Federated weights",
"mounted_path": "/weights/openfold3/fed"
},
{
"model_type": "openfold3",
"version": "3.0.0-fine-tuned",
"model_version_id": "openfold3:1.2.30",
"model_version_tag": "1.2.3-openfold3-by-file",
"description": "OpenFold Fine-Tuned weights",
"mounted_path": "/weights/openfold3/fine-tuned"
}
]
}
The Deployment Examples below illustrate how to set up each method in both Docker and Kubernetes environments. OpenFold3 is used in the examples, but the same principles apply to Boltz-2.
Kubernetes Deployment🔗
This section provides detailed steps for deploying custom weights in a Kubernetes environment using either configuration method.
Important
Ensure your weight files are organized following the structure shown in Required Files and stored in a location accessible by your Kubernetes cluster (e.g., PersistentVolumeClaim backed by EFS, NFS, or hostPath for testing).
Configure Helm Values🔗
Choose your configuration method:
Method 1: Configuration File🔗
Create a ConfigMap with your weights configuration:
apiVersion: v1
kind: ConfigMap
metadata:
name: openfold3-weights-config
namespace: apheris-hub
data:
weights.json: |
{
"available_weights": [
{
"model_type": "openfold3",
"version": "3.0.0-federated",
"description": "OpenFold Federated weights",
"mounted_path": "/weights/openfold3/fed"
},
{
"model_type": "openfold3",
"version": "3.0.0-fine-tuned",
"description": "OpenFold Fine-Tuned weights",
"mounted_path": "/weights/openfold3/fine-tuned"
}
]
}
Apply the ConfigMap:
kubectl apply -f weights-config.yaml
Update your Helm values.yaml:
models:
instances:
- name: openfold3
image: quay.io/apheris/hub-apps:1.2.3-openfold3-by-file
env:
- name: APH_WEIGHTS_CONFIG_FILE
value: /config/weights.json
extraVolumes:
- name: weights-storage
persistentVolumeClaim:
claimName: custom-weights-pvc
- name: weights-config
configMap:
name: openfold3-weights-config
extraVolumeMounts:
- name: weights-storage
mountPath: /weights
readOnly: true
- name: weights-config
mountPath: /config
readOnly: true
Method 2: Environment Variable🔗
Update your Helm values.yaml:
models:
instances:
- name: openfold3
image: quay.io/apheris/hub-apps:1.2.3-openfold3-by-file
env:
- name: APH_AVAILABLE_WEIGHTS
value: |
[
{
"model_type": "openfold3",
"version": "3.0.0-federated",
"description": "OpenFold Federated weights",
"mounted_path": "/weights/openfold3/fed"
},
{
"model_type": "openfold3",
"version": "3.0.0-fine-tuned",
"description": "OpenFold Fine-Tuned weights",
"mounted_path": "/weights/openfold3/fine-tuned"
}
]
extraVolumes:
- name: weights-storage
persistentVolumeClaim:
claimName: custom-weights-pvc
extraVolumeMounts:
- name: weights-storage
mountPath: /weights
readOnly: true
JSON Formatting in YAML
When using the environment variable method, ensure your JSON is properly formatted within the YAML value: | block. Each line should be indented consistently.
Deploy with Helm🔗
Upgrade your Helm release with helm upgrade command.
Pod Restart Required
If you're adding custom weights to an existing deployment, you must restart the model pod for changes to take effect, for example for OpenFold3 use:
kubectl rollout restart deployment/hub-openfold3 -n apheris-hub
Verify Configuration🔗
Verify Environment Variables🔗
kubectl exec -n apheris-hub deployment/hub-openfold3 -- env | grep APH_
You should see either APH_WEIGHTS_CONFIG_FILE or APH_AVAILABLE_WEIGHTS set.
Check Volume Mounts🔗
kubectl exec -n apheris-hub deployment/hub-openfold3 -- ls -la /weights/openfold3/
Verify your weight directories are present:
drwxr-xr-x 2 root root 4096 Jan 15 10:00 fed
drwxr-xr-x 2 root root 4096 Jan 15 10:00 fine-tuned
Test Weights Endpoint🔗
Port-forward to the model service:
kubectl port-forward -n apheris-hub svc/hub-openfold3 8000:8000
Query the weights endpoint:
curl http://localhost:8000/weights
Expected response:
{
"available_weights": [
...
{
"model_type": "openfold3",
"version": "3.0.0-federated",
"description": "OpenFold Federated weights",
"mounted_path": "/weights/openfold3/fed"
},
{
"model_type": "openfold3",
"version": "3.0.0-fine-tuned",
"description": "OpenFold Fine-Tuned weights",
"mounted_path": "/weights/openfold3/fine-tuned"
}
]
}
Docker Deployment🔗
This section provides examples for running custom weights with Docker using either configuration method.
Method 1: Configuration File (Docker Volume Mount)🔗
Prepare your directory structure:
mkdir -p weights_mount/openfold3/fed
mkdir -p weights_mount/openfold3/fine-tuned
# Copy your weight files
cp -r /path/to/federated/weights/* weights_mount/openfold3/fed/
cp -r /path/to/fine-tuned/weights/* weights_mount/openfold3/fine-tuned/
Create configuration file weights_mount/additional_weights.json:
{
"available_weights": [
{
"model_type": "openfold3",
"version": "3.0.0-federated",
"description": "OpenFold Federated weights",
"mounted_path": "/weights/openfold3/fed"
},
{
"model_type": "openfold3",
"version": "3.0.0-fine-tuned",
"description": "OpenFold Fine-Tuned weights",
"mounted_path": "/weights/openfold3/fine-tuned"
}
]
}
Run a Docker container setting the APH_WEIGHTS_CONFIG_FILE environment variable and mounting the weights and configuration directory:
docker run --name openfold3 \
-v $(pwd)/weights_mount:/weights/openfold3 \
-e APH_WEIGHTS_CONFIG_FILE=/weights/openfold3/additional_weights.json \
...
Path References
The mounted_path in the JSON file refers to paths inside the Docker container, not on the host system. Please make sure that both the weights and configuration file are correctly mounted into the container.
Method 2: Using Environment Variable (Docker Inline JSON)🔗
Prepare your directory structure (same as Method 1):
mkdir -p weights_mount/openfold3/fed
mkdir -p weights_mount/openfold3/fine-tuned
# Copy your weight files
cp -r /path/to/federated/weights/* weights_mount/openfold3/fed/
cp -r /path/to/fine-tuned/weights/* weights_mount/openfold3/fine-tuned/
Run Docker container with inline JSON:
docker run --name openfold3 \
-v $(pwd)/weights_mount:/weights/openfold3 \
-e APH_AVAILABLE_WEIGHTS='[
{
"model_type": "openfold3",
"version": "3.0.0-federated",
"description": "OpenFold Federated weights",
"mounted_path": "/weights/openfold3/fed"
},
{
"model_type": "openfold3",
"version": "3.0.0-fine-tuned",
"description": "OpenFold Fine-Tuned weights",
"mounted_path": "/weights/openfold3/fine-tuned"
}
]' \
...
Verify Docker Deployment🔗
Verify environment variables🔗
docker exec openfold3 env | grep APH_
Check mounted weights🔗
docker exec openfold3 ls -la /weights/openfold3/
Test weights endpoint🔗
curl http://localhost:8000/weights
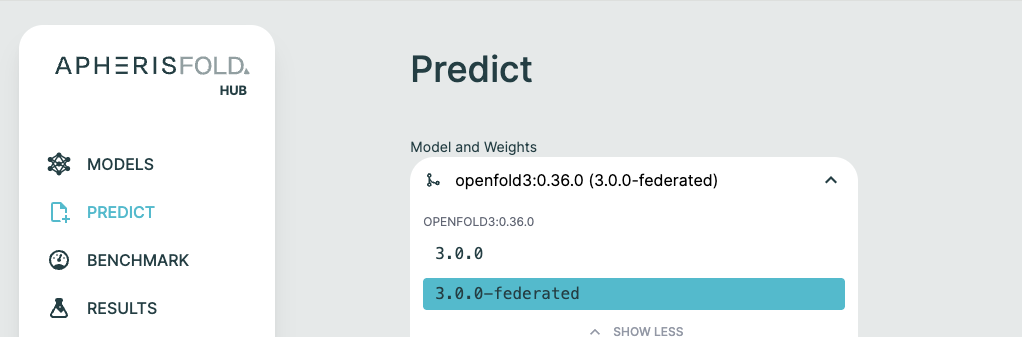
Using Custom Weights🔗
Once configured, custom weights appear in the Hub UI alongside default weights. When creating a prediction:
- Navigate to the Predict page
- Select your model (OpenFold3 or Boltz-2)
- Select your custom weight version from the dropdown
When using the API, specify the custom weight version in your prediction payload:
{
"id": "1",
"inputPath": "input/assets",
"outputPath": "output/results",
"requestParams": {"queries": {...}},
"modelName": "openfold3",
"weightVersion": "3.0.0-federated"
}
Multiple Sequence Alignment (MSA)🔗
MSAs for ApherisFold can be provided either via pre-computed .a3m files, or by using a compatible MSA server implementation.
The default behavior for models is to use user-supplied pre-generated MSA (.a3m) files with an option to use an existing MSA server.
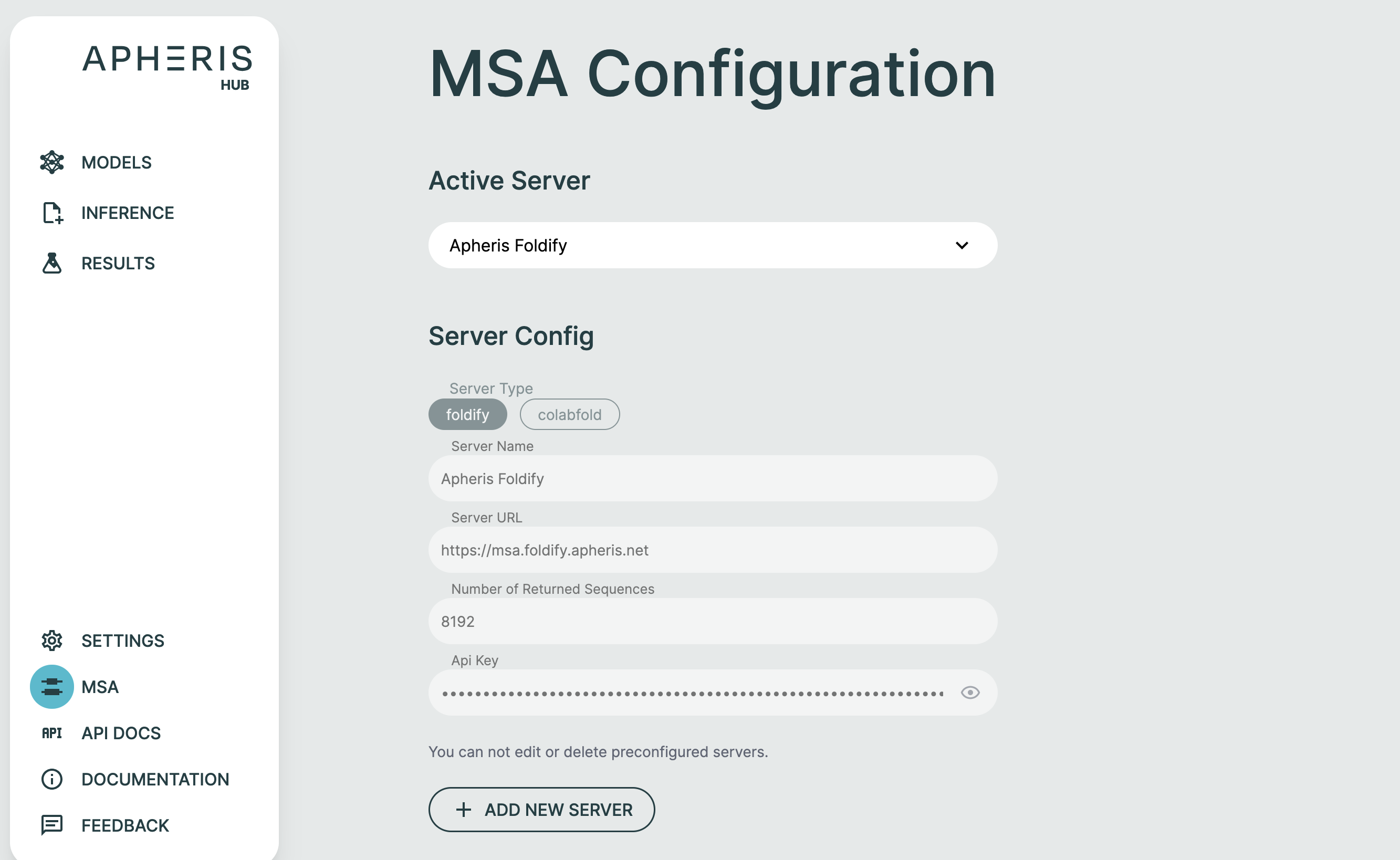
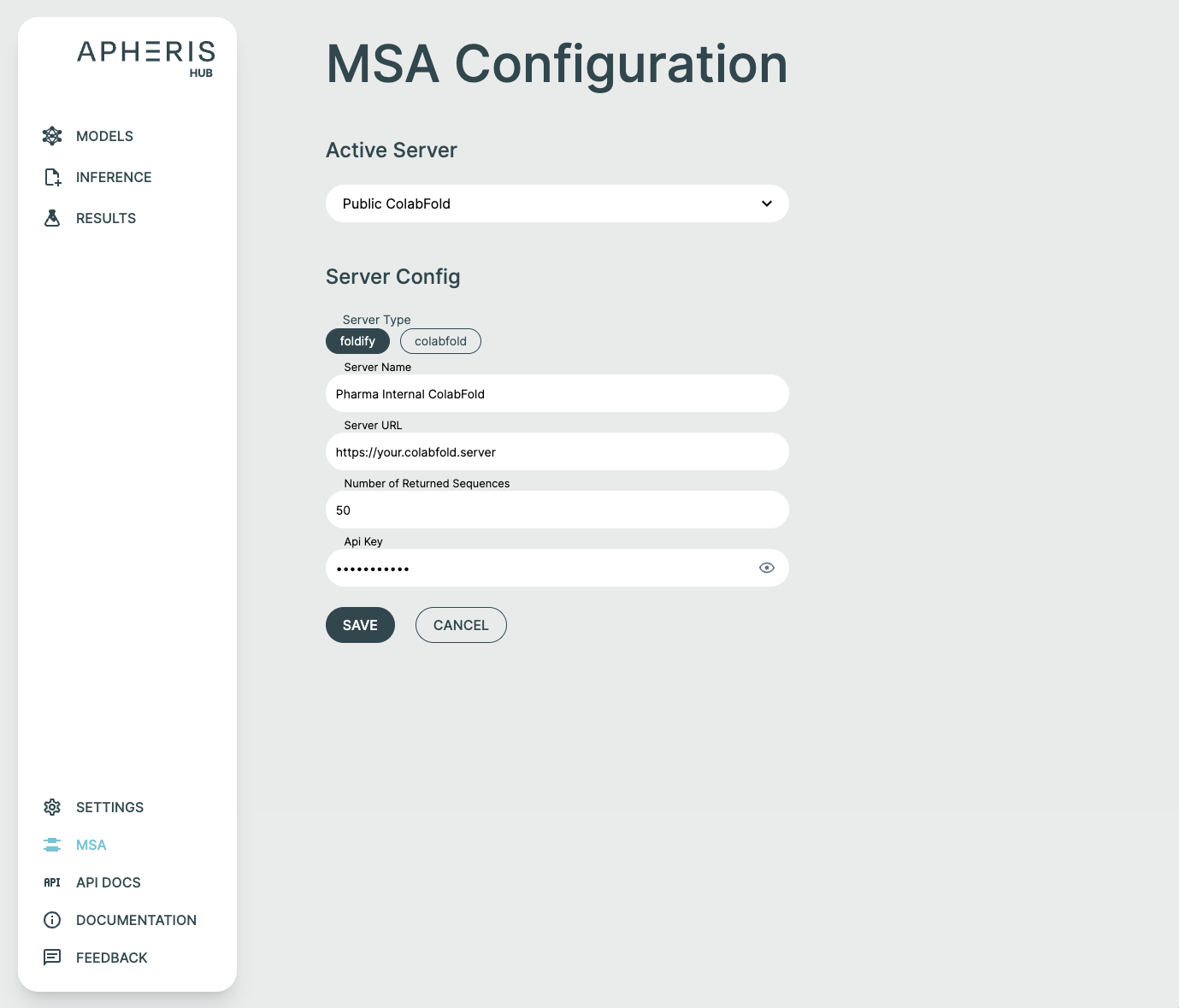
ApherisFold supports ColabFold, NVIDIA NIM ColabFold, and Foldify servers, and these can be configured using the MSA page in the Apheris Hub. From this page you can specify the type of server, its URL and, in the case of Foldify, the API key.
For a limited time the Hub ships with free evaluation access to an Apheris-hosted MSA server that can optionally be used. The Apheris-hosted public MSA server should be used only for evaluation and be treated as a public MSA API. If you'd like assistance with deploying your own MSA server, please contact us.
Important
Any queries submitted using the Apheris-hosted Foldify server or public ColabFold versions of models will be sent to the MSA server hosted by, at least, ColabFold, Foldify or Apheris. If you do not wish to have your sequences leave your environment, use the default model and supply your pre-generated MSA files or use a self-deployed ColabFold or Foldify server.
Configurable MSA Server🔗
You can easily add your own ColabFold, NVIDIA NIM ColabFold, or Foldify MSA server via the MSA option.
To configure your server simply supply:
- Server Name
- Server Type: ColabFold, NVIDIA NIM ColabFold, or Foldify
- Server URL
- Number of Returned Sequences
- API Key
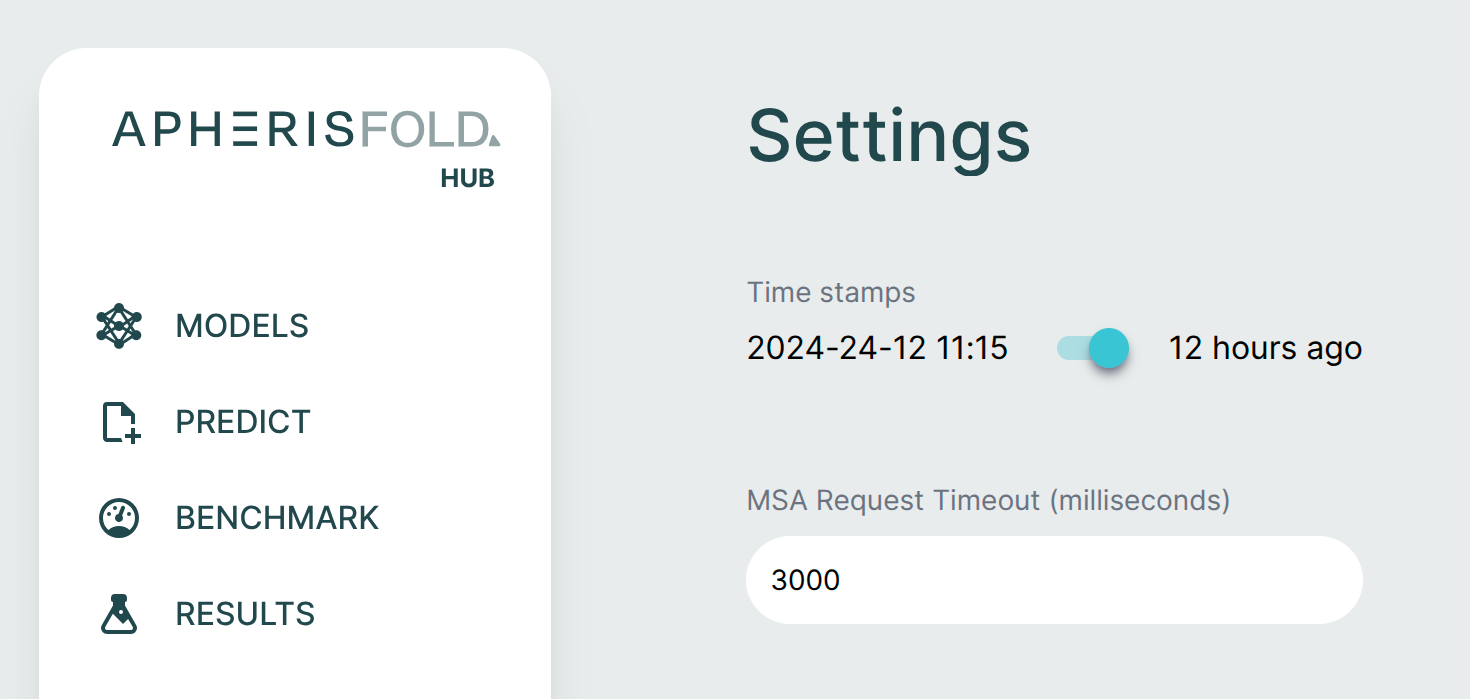
MSA Request Timeout (ColabFold Only)🔗
For ColabFold servers, you can configure the timeout for MSA server requests in the Settings page. This setting controls how long the browser will wait for a response from your ColabFold server before timing out. This timeout setting only applies to requests made directly from your browser to the MSA server, it does not affect backend requests or model prediction timeouts.
To adjust the timeout:
- Navigate to the Settings page in the Apheris Hub
- Locate the MSA Request Timeout field
- Enter your desired timeout value in milliseconds
- The default timeout is 30000ms (30 seconds)
MSA Usage🔗
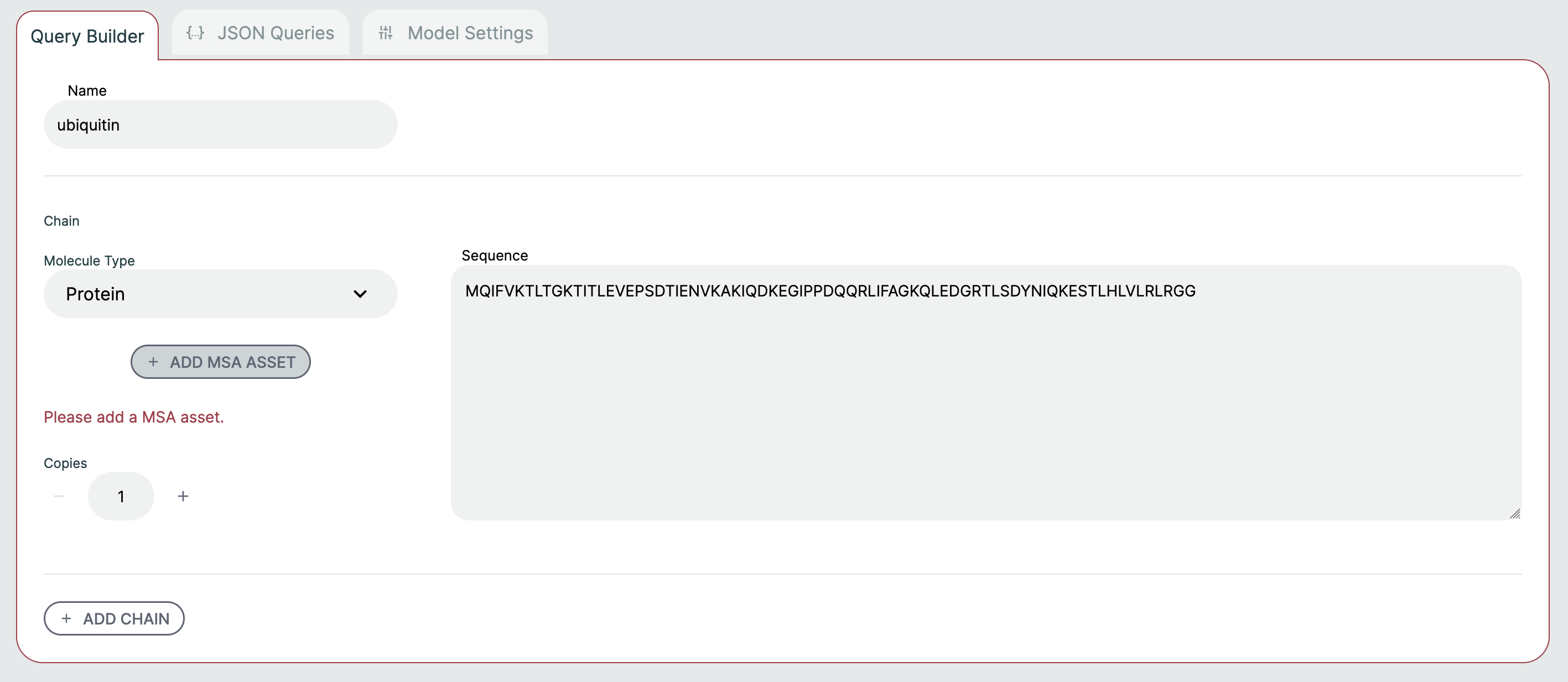
Unless an MSA server is specifically enabled, ApherisFold will expect you to provide MSAs for protein sequences by uploading an a3m file. You can do this by clicking the "ADD MSA ASSET" button on the Query Builder, then selecting the .a3m from your file system. If using this method, the ApherisFold application will not reach out to any external servers. For each protein chain in the request, add an extra "msa": "filename.a3m" field (input validation will fail if the msa field is missing).
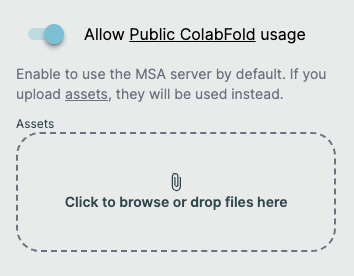
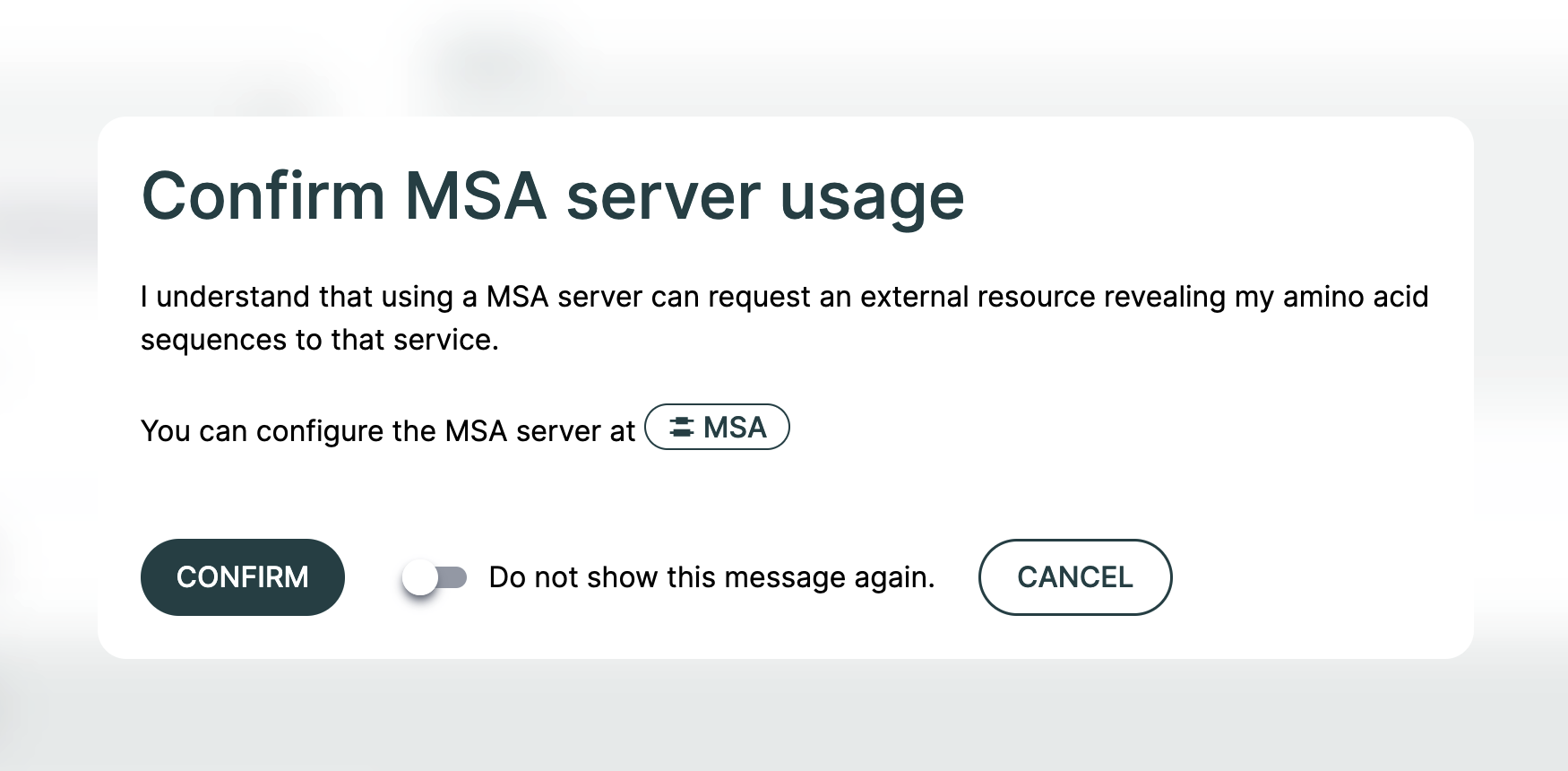
To enable an MSA server for your query, use the switch below the Query Builder entitled "Allow [MSA Server Name] usage". When activating this, you will receive a warning stating that your sequences will be processed by an external service. If accepted, this will not show again for the duration of your browser session.
Enabling this option will generate the MSAs for the request at prediction time and ignore any supplied MSA files. You will also receive a prompt to confirm you want to do this and advise that doing so may result in sending requests to an external server.
See the Running Prediction section for an example of using your own MSA files for prediction.
Apheris can also help you set up a private MSA server. In that case, please contact us.
Running Prediction🔗
Running prediction can be done via the UI or the Hub API. Here, we cover running prediction using the UI.
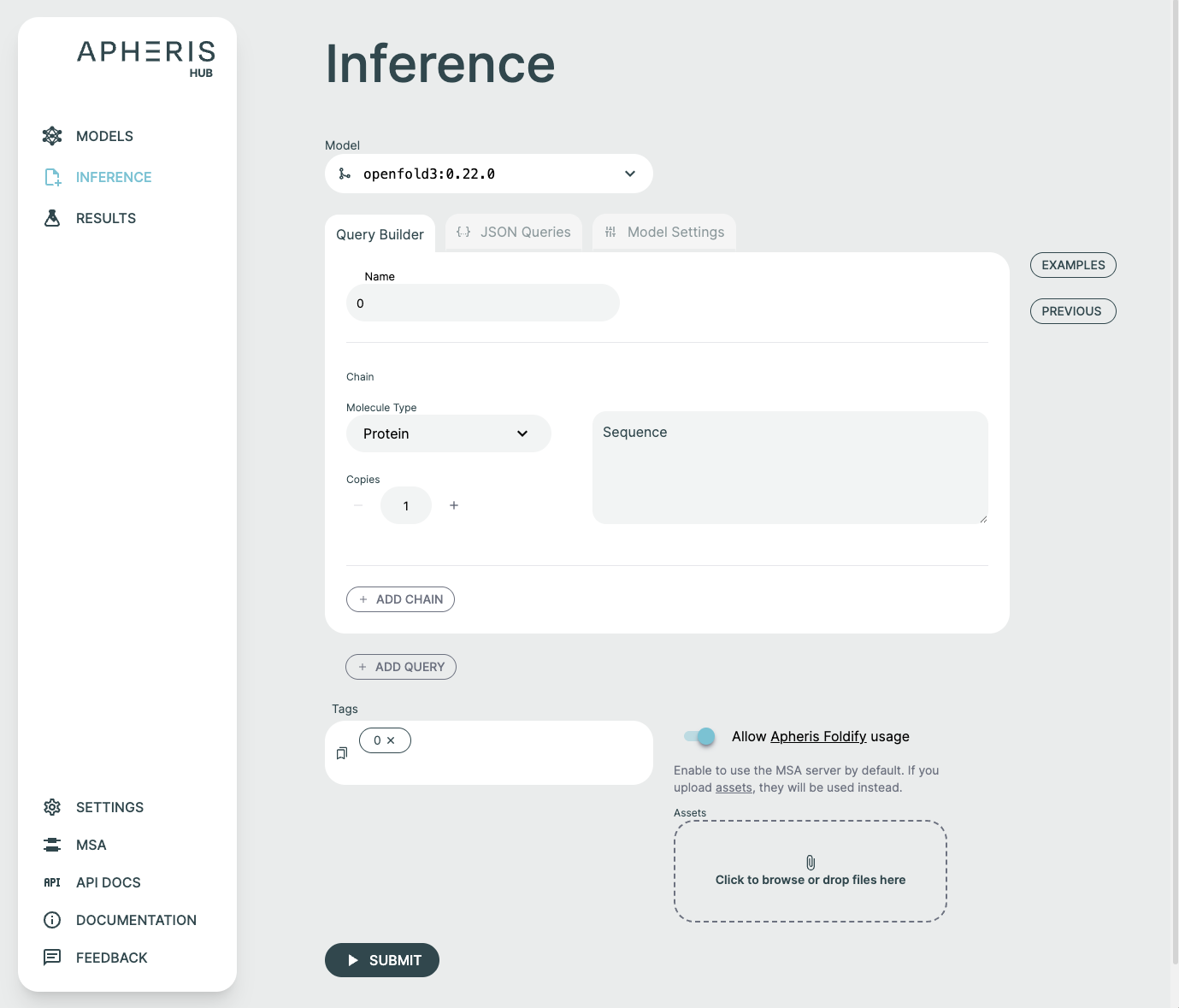
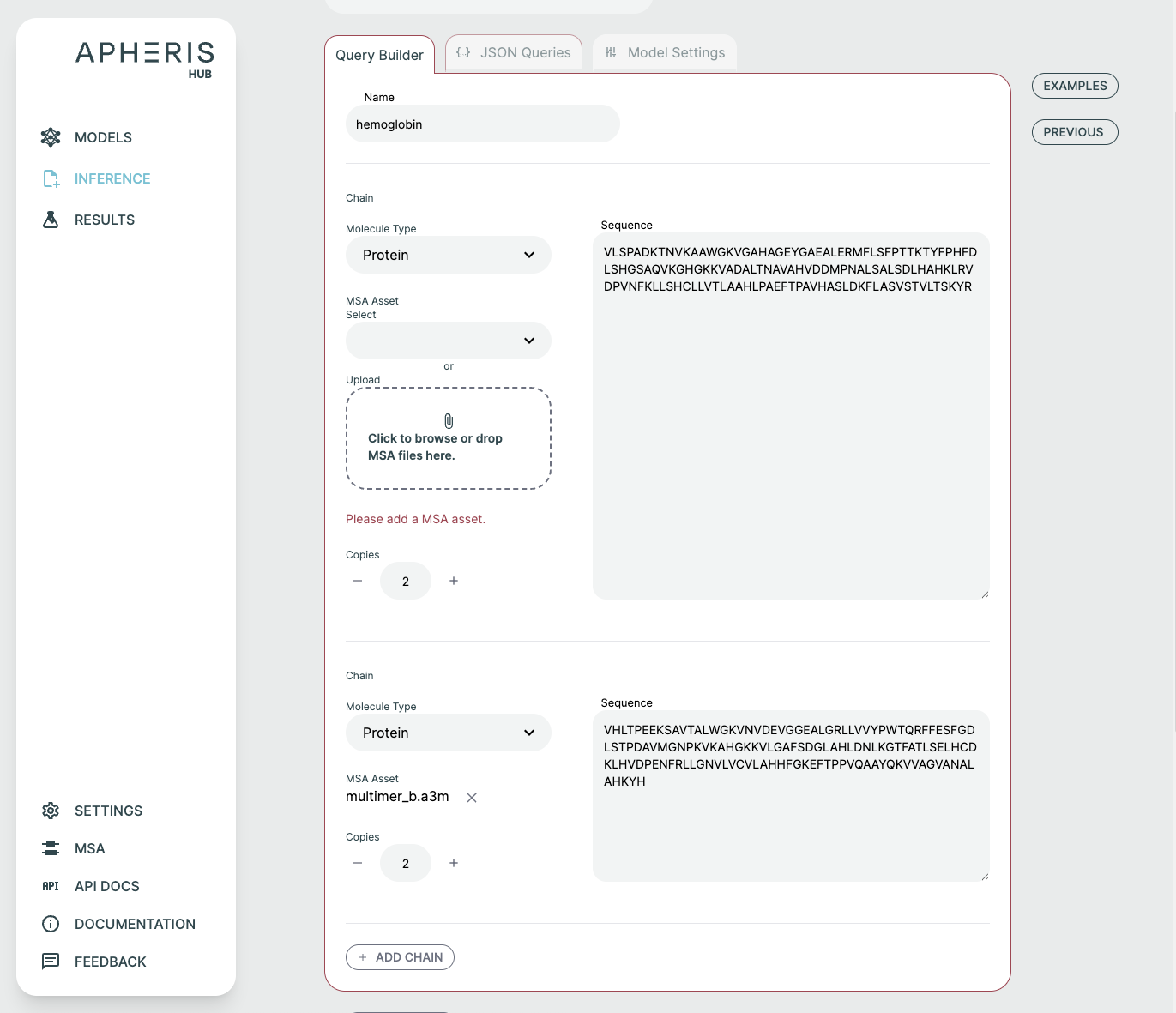
Query Builder🔗
It is possible to form a prediction query using the graphical Query Builder and the JSON Queries Editor.
You can add to and adjust the query to fit your needs. It is possible to make changes as well as add additional components or chains to the query.
Here are some notable ways to configure a query:
- Molecule Type: Select between Protein, Ligand, RNA, and DNA. There are some type-specific features such as the ability to use SMILES vs CCD code for ligands. You can specify multiple ligands either through separate components with different identifiers, or by separating them with
.as part of one SMILES string, e.g.CCCC.CO. Note that this approach will not result in successful comparisons to attached ground-truth structural data, since this relies on each ligand being assigned a unique chain identifier in both the predicted and attached structures. - Copies: Adjusts the number of copies for the chain. A name is automatically created and usually does not need to be modified. If you'd like to adjust the name, this can be done in the JSON Queries tab.
- Add Chain: Adds a chain to the query. If you need to delete the chain, hover the mouse to the top right. There must always be at least one chain per query.
- Add Query: Add a new query to the request. Hover the mouse to the top right of the query to delete it. Delete will not appear if there is only one query as there must always be at least one query.
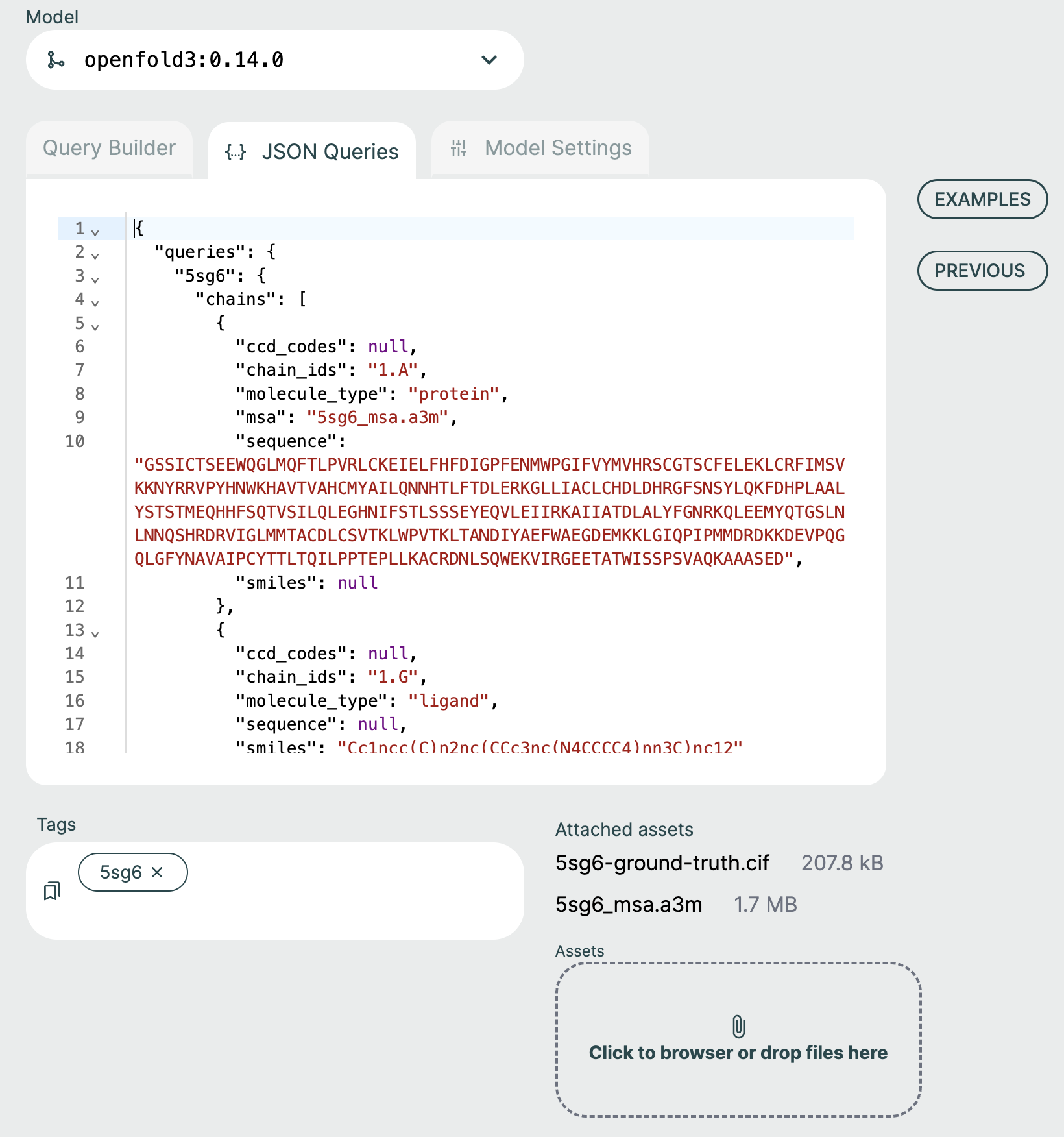
JSON Queries Editor🔗
The JSON Queries Editor view is considered an advanced view and it is recommended to instead use the Query Builder to form new queries. The queries made via the Query Builder are reflected in the JSON Editor view but not all changes in the JSON Editor will be reflected back to the Query Builder.
For advanced users, the JSON Editor offers many convenience features such as syntax highlighting, keyword auto-completion, and data type validation.
Tags🔗
Query names that are supplied in the JSON payload are automatically derived as tags for the request. These tags can later be used to search for specific prediction requests.
Custom Tags🔗
You can supply your own tags by typing directly in the "Tags" field. These tags will appear with the results and can be searched for fast lookups across all queries and results. Custom tags are a great way to distinguish between requests that might have similar queries.
Examples🔗
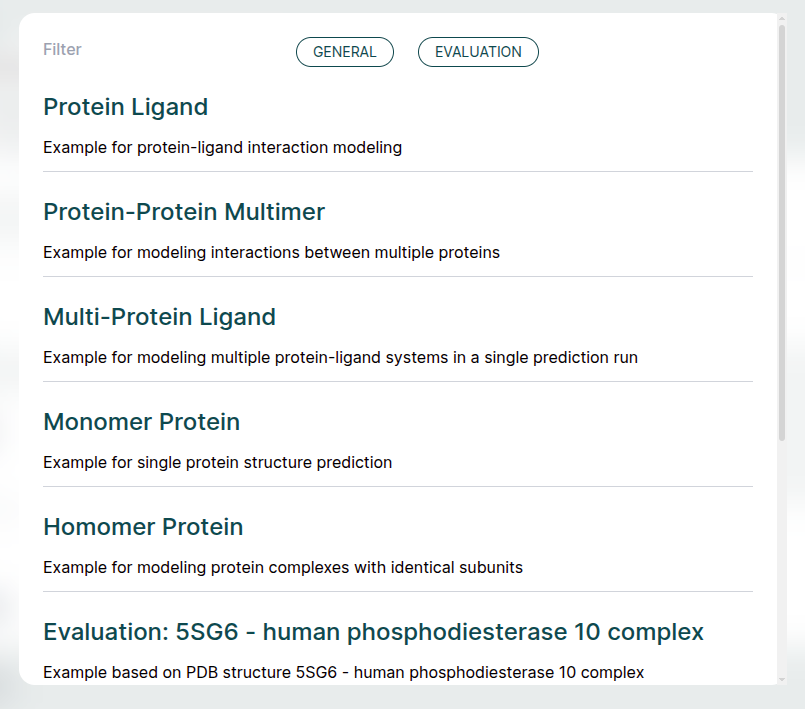
The ApherisFold application comes with examples to help with getting started. Each of these examples can be run across all models. These can be used as starting templates for your queries or as a simple way to evaluate the full workflow.
The examples provide MSA files and do not need a MSA server to run. The examples can also be used with configured MSA servers, as described in Multiple Sequence Alignment (MSA).
Below we provide a bit more detail on each example:
- Protein Ligand: The simple use-case of co-folding a small molecule with a single protein chain. The protein chain corresponds to the sequence of MCL1, and the small molecule is Aceclidine.
- Protein-Protein Multimer: Hemoglobin is an example of a query consisting of a multimer of protein chains, with no ligand present.
- Multi-Protein Ligand: A single query consisting of multiple sub-queries. In this case, MCL1 is co-folded with two different ligands (Aceclidine and 1-benzothiophene-2-carboxylic acid).
- Monomer Protein: A single protein chain, Ubiquitin.
- Homomer Protein: A dimer of identical protein chains, in this case the GCN4 Leucine Zipper.
Evaluation Examples🔗
The remaining examples illustrate how the ApherisFold platform can be used to evaluate models on reference structures. Each one is identified by a PDB ID (since the data in these examples is derived from the public domain) and is accompanied by a "{query_name}-ground-truth.cif" file attached as an asset.
Note
If you provide your own .cif files to serve as ground-truth, the name of the file should match the name of the query in your request.
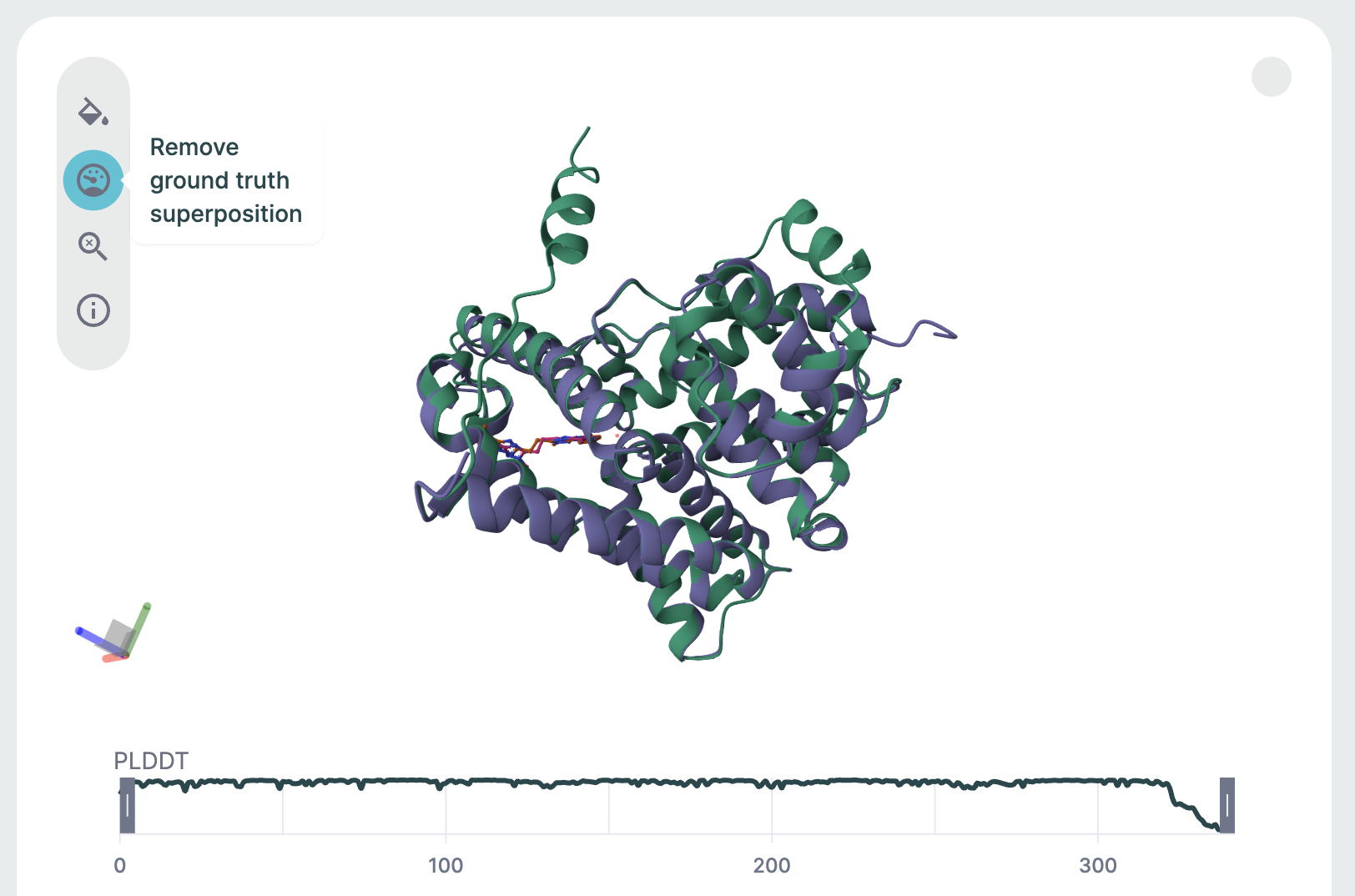
After predicting the structure, the prediction is aligned to this reference structure, and several metrics of structural similarity are computed. The superimposed structures and metrics can then be viewed in the corresponding results page.
Assets and MSA files🔗
If your request requires additional assets, such as an MSA file, they can be supplied by clicking the Assets button or dropping in files.
Once you have uploaded your assets, they need to be included in your query by clicking the MSA Asset selection dropdown to choose from already added files or upload a new file. The JSON Editor will reflect this with the msa field.
If you want to set your request up as an evaluation of model performance on a known structure, make sure to upload the reference CIF file with the naming schema "{query_name}-ground-truth.cif".
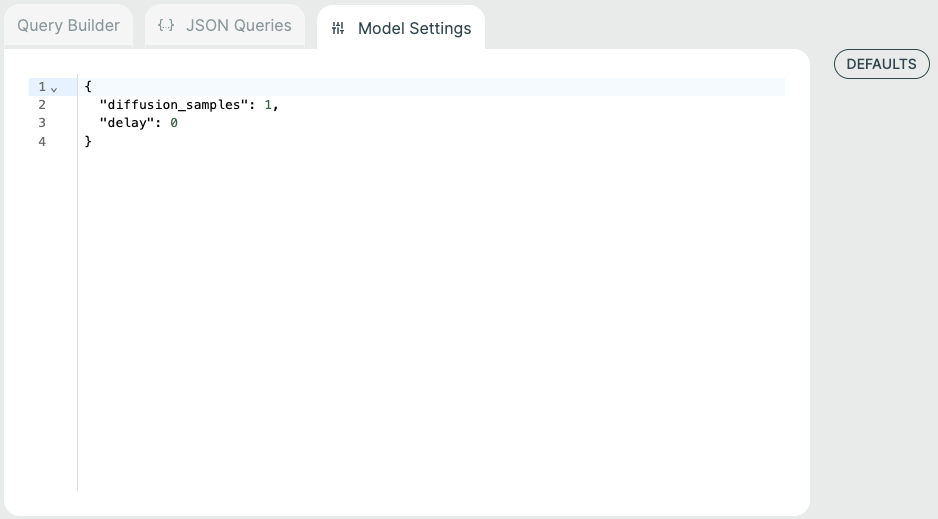
Model Settings🔗
Some models have additional model-specific settings that can be configured such as diffusion samples. These can be changed on the Prediction screen by selecting the "Model Settings" tab at the top.
You can click the "Defaults" button to see the available model settings and make changes.
Results Management🔗
Whatever you submit in the JSON payload (one or multiple queries) is submitted as a single request.
Submitting additional requests will place those requests in a queue. The GPU is fully consumed per request. Parallelism and multi-GPU support will come in follow-up releases.
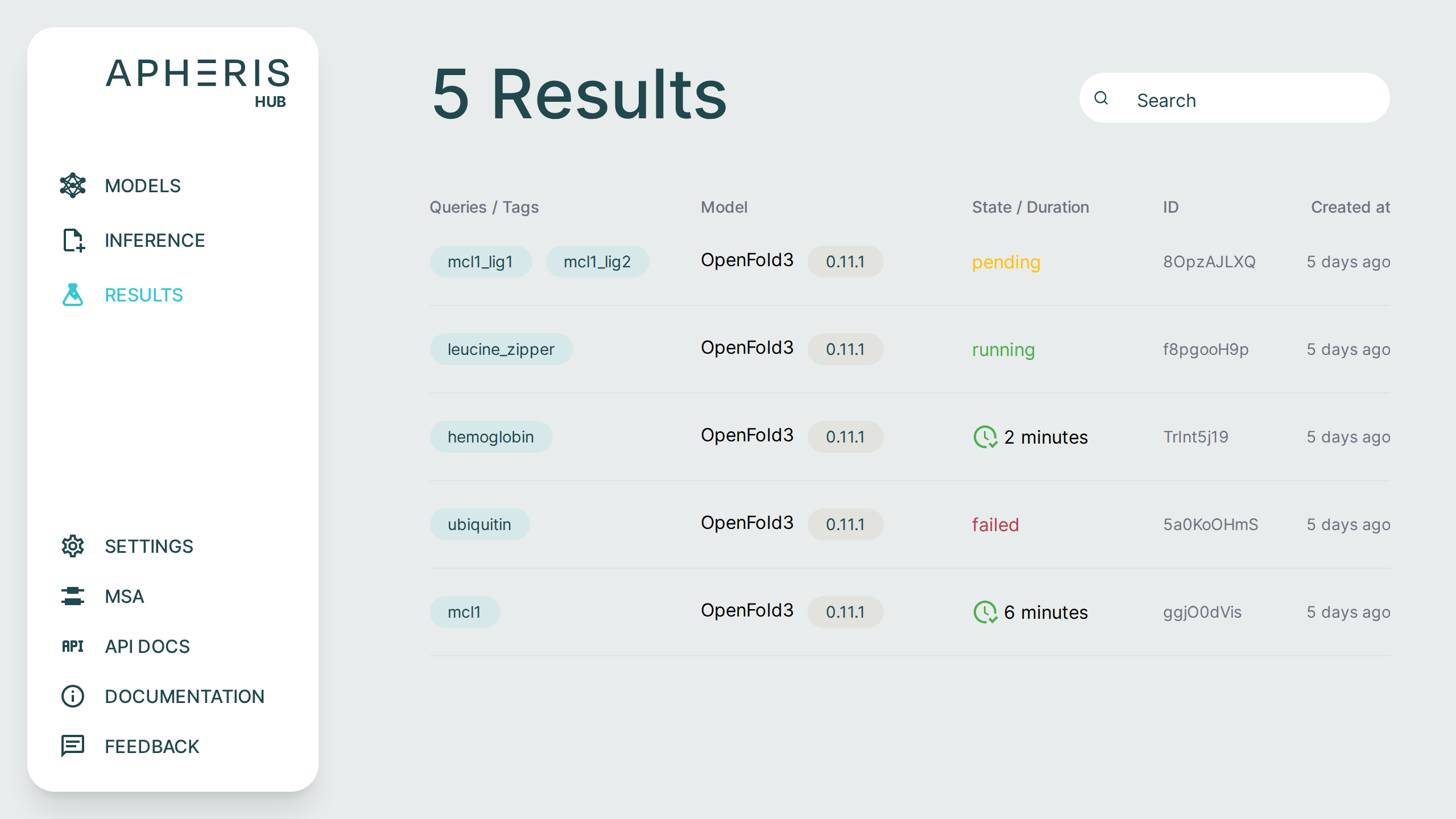
All submitted requests and results can be viewed on the Results page.
Request Search🔗
You can also filter down results based on tags set for the queries or for metadata such as model name.
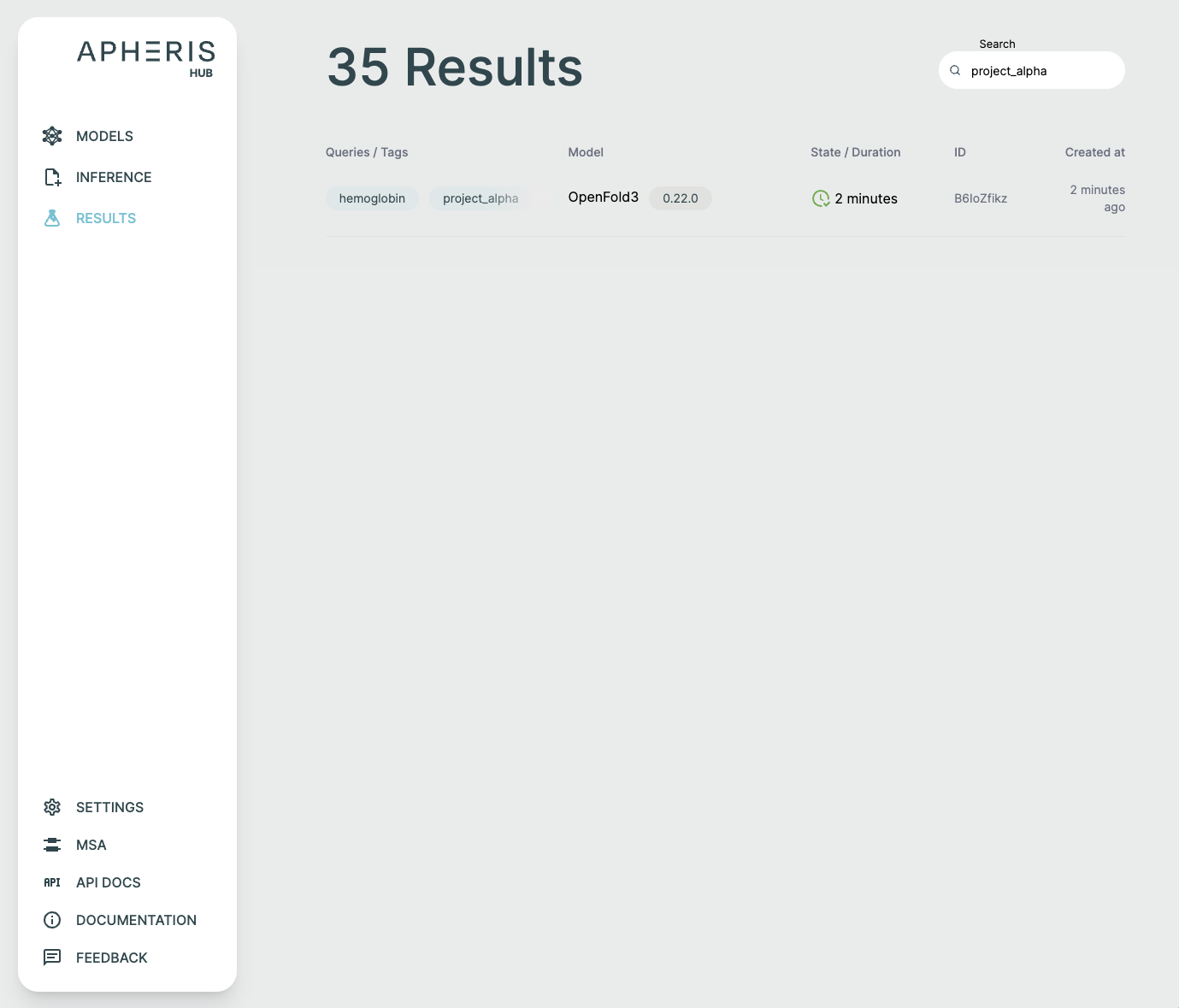
Request Status🔗
Submitted requests can have a few statuses:
- Pending
- Running
- Failed
- Done
- When completed, the status shows the total query runtime.
In addition to the request status, you can also see when the request was created, the model and version, and any tags associated with the request.
Reuse a Request in a New Prediction🔗
On any Results detail page, the Job Details section includes a Set up new prediction button. Selecting it opens the Prediction page with the original request pre-populated:
- Request parameters load into the Query Builder and JSON editor in the same format the API expects.
- Tags, model version, and model settings are restored when available.
All fields remain editable and submitting creates a brand-new request, leaving the original results unchanged.
Results Deletion🔗
Request results are persisted in the output folder specified when you deployed the Apheris Hub for the first time, either via Docker or Kubernetes.
Currently, there is no built-in way to delete these results. Instead, they are managed through the file system. This is an intentional measure to avoid any accidental deletion of valuable insights.
Analyzing Results🔗
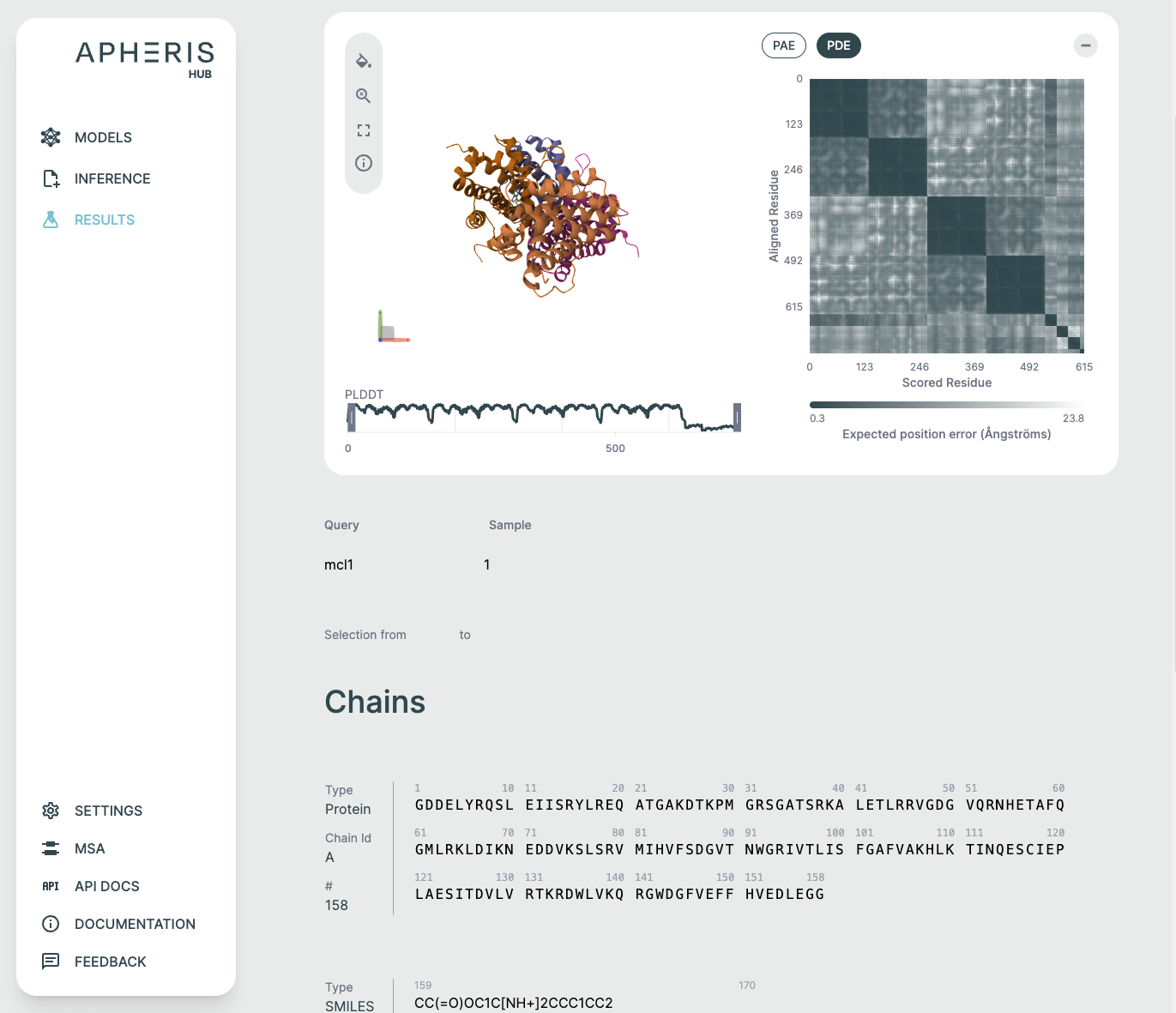
The ApherisFold application comes pre-packaged with specialized analysis tools to support analyzing prediction results within the UI.
You can also Download Results by clicking the raw results download button at the top of the results screen.
Each of these visual components serves a specific purpose in interpreting co-folding predictions:
- 3D viewer: Understand overall fold and domain organization
- PAE/PDE: Assess inter-residue or inter-chain confidence
- pLDDT plot: Gauge per-residue reliability
- Sequence section: Review input-output fidelity and interpret ligand participation
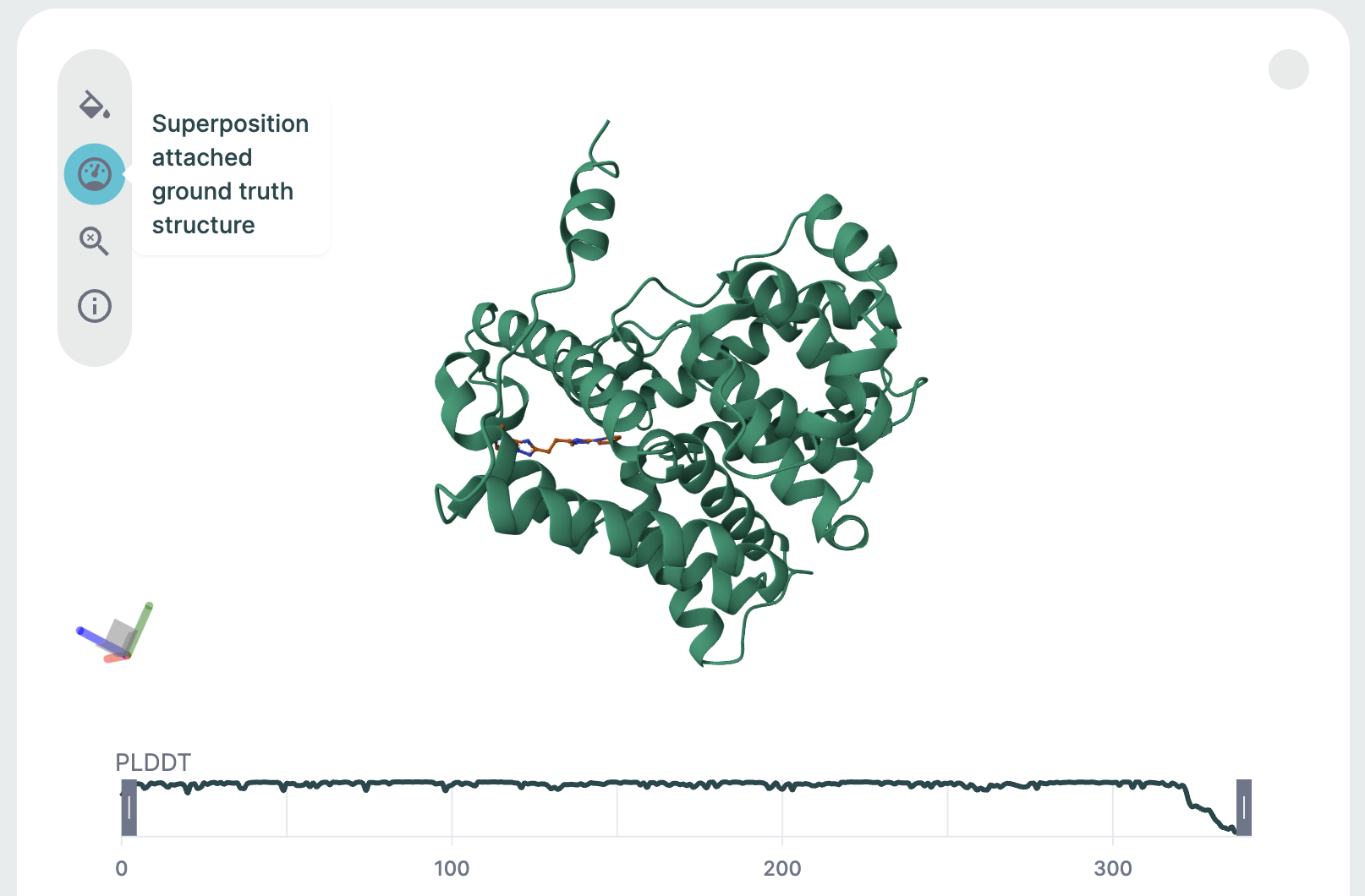
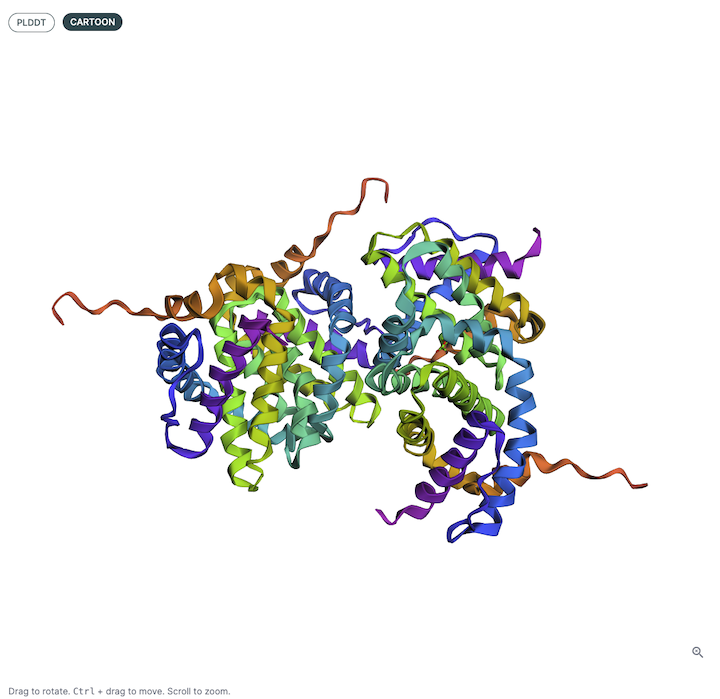
3D Structure Viewer (Left Panel)🔗
This panel displays the 3D atomic structure of the predicted protein or protein-ligand complex. The cartoon representation emphasizes the secondary structure elements (helices, sheets, loops).
Viewer Color Coding🔗
The structure is colored based on per-residue pLDDT scores, which indicate the model’s confidence in the predicted atomic positions. The scale typically follows:
- Blue: Very high confidence (pLDDT > 90)
- Green: Confident (70–90)
- Yellow/Orange: Low confidence (50–70)
- Red: Very low confidence (< 50)
This can be helpful for visualizing local and global structure quality, identifying disordered or uncertain regions, and verifying expected fold topology.
PAE / PDE Matrix (Right Panel)🔗
The Predicted Aligned Error (PAE) or Predicted Distance Error (PDE) matrices visualize the model's expected error in positioning residue pairs relative to one another. This is essential for interpreting inter-domain and inter-chain interaction confidence and is especially relevant in multi-chain or protein-ligand complexes.
- PDE: (specific to Boltz-2) is a measure of the uncertainty of the model in the distance between two residues/ligand atoms in the prediction, which is useful for uncertainty-aware modeling.
- PAE: further captures the uncertainty of the model in the relative orientation, in addition to the distance, of pairs of residues/ligand atoms in the prediction. For further reading, there is an excellent visualization created by the European Molecular Biology Laboratory (EMBL) on this guide.
Both of these are most easily visualized as a heat map, where each axis corresponds to sequential Amino Acid/DNA/RNA residues or ligand atoms.
Matrix Color Coding🔗
Darker cells imply lower predicted error (higher confidence), while lighter regions suggest areas of structural uncertainty.
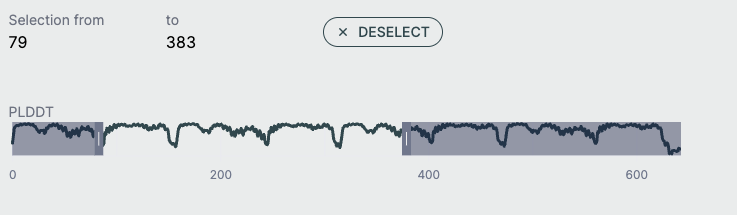
pLDDT Plot🔗
This line graph shows the pLDDT score per residue, providing a quick global overview of prediction confidence. It's ideal for identifying low-confidence loops or disordered regions and useful for downstream filtering (e.g., in docking or dynamics simulations).
- X-axis: Residue index across all chains
- Y-axis: pLDDT score (0–100)
Full-Screen Molecular Viewer🔗
The molecular viewer can be maximized to occupy most of the screen space for detailed structural inspection. In full-screen mode, the 3D viewer, PAE/PDE plot, and sequence bar are displayed together to provide a comprehensive view of the model. Each component can be minimized or expanded individually as needed, while the 3D viewer remains as the main background element for structure visualization.
See the video below for a demonstration of the full-screen view.
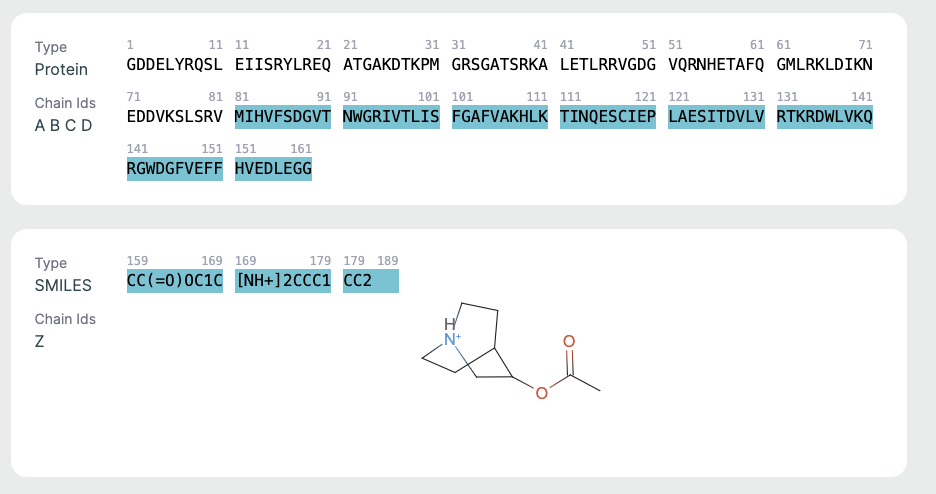
Chains Sequence Display🔗
This part is helpful for verifying sequence input/output consistency. Ligand info is key for those focused on drug binding or active site modeling.
- Displays the full amino acid sequence used for the prediction.
- Includes chain identifiers (A, B, C, etc.).
Ligand Representation (SMILES)🔗
If a ligand is present (as in co-folding), it is shown in SMILES format along with a 2D molecular rendering.
Guide: Benchmarking Using ApherisFold🔗
In a future release, ApherisFold will support interactive benchmarking directly within the Apheris Hub UI, allowing you to compare models and outputs side by side.
In the meantime, you can perform benchmarking on your data programmatically using the Apheris Hub API. This guide walks you through benchmarking OpenFold3 models, capturing metrics, and recording them locally using a short Python script. If you wish, you can adapt the approach to any language that supports HTTP requests.
Querying OpenFold3 Using the Hub API🔗
Before we begin, here's a small helper function for pretty-printing API responses:
import json
import requests
def pretty_print_dict(response: dict) -> None:
print(json.dumps(response, indent=2))
Verifying Installed Models🔗
To check which models and versions the Hub has discovered and marked as available, run:
HUB_API_URL = "https://replace-me-with-your-url/api/v1/"
response = requests.get(HUB_API_URL + "models/installed")
pretty_print_dict(response.json())
Example response:
{
"data": {
"openfold3": {
"versions": {
"openfold3:0.28.0": {
"status": "available",
"endpoint": "http://openfold3-service:8000",
"weights": [
{
"version": "3.0.0",
"description": "Default OpenFold3 weights"
}
]
}
}
}
}
}
Success: OpenFold3 has been discovered and is available via the Hub.
Let’s move on to preparing input data.
Preparing Inputs🔗
To perform prediction using ApherisFold via the Hub API, create JSON files specifying the protein sequences, ligand identifiers (SMILES), or cofactors/DNA/RNA to co-fold.
Example schemas can be found in the Apheris Hub documentation.
You can also generate queries directly from CIF files (PDB/mmCIF) to benchmark OpenFold3. Use the endpoint returned under endpoint in the previous API response:
payload = {
"input_path": "CIF_FILE_DIRECTORY",
"cif_file_names": ["5sfc", "5shh"],
}
response = requests.post(HUB_API_URL + "/query_from_mmcif", json=payload)
pretty_print_dict(response.json())
Example response (truncated for brevity):
{
"queries": {
"5sfc": {
"chains": [
{ "molecule_type": "protein", "chain_ids": "1.A", "sequence": "..." },
{ "molecule_type": "ligand", "chain_ids": "1.G", "smiles": "..." }
]
},
"5shh": { ... }
},
"errors": {}
}
Container Volume Structure🔗
The ApherisFold container ingests files through a mounted host volume (/apheris/input). Your CIF directory must be located within this mount point. For example:
/apheris/input/
├── cif
│ └── directory
│ ├── 5sfc.cif
│ ├── 5shh.cif
If errors occur, failing file names will appear in the errors field with details. Each successfully extracted query appears under queries.
Adding Multiple Sequence Alignments (MSAs)🔗
OpenFold3 can run without MSAs, but predictions will be of lower quality. You’ll need to map protein sequences to MSA filenames and inject them into each query:
query_dict = response.json()["queries"]
for query_name, query in query_dict.items():
for chain in query["chains"]:
if chain["molecule_type"] == "protein":
chain["msa"] = PROTEIN_SEQUENCE_TO_MSA_NAME[chain["sequence"]]
Building the Prediction Payload🔗
Now, define your benchmark and construct the full payload:
YOUR_BENCHMARK_NAME = "example-benchmark"
payload = {
"input_path": "CIF_FILE_DIRECTORY",
"cif_file_names": ["5sfc", "5shh"],
}
response = requests.post(HUB_API_URL + "/query_from_mmcif", json=payload)
query_dict = response.json()["queries"]
for query_name, query in query_dict.items():
for chain in query["chains"]:
if chain["molecule_type"] == "protein":
chain["msa"] = PROTEIN_SEQUENCE_TO_MSA_NAME[chain["sequence"]]
prediction_payload = {
"id": "1",
"inputPath": f"{YOUR_BENCHMARK_NAME}/assets",
"outputPath": f"{YOUR_BENCHMARK_NAME}/outputs",
"requestParams": {"queries": query_dict},
"modelParams": {"diffusion_samples": 1},
"modelName": "openfold3",
"modelVersion": "3.0.0"
}
Adding Ground-Truth Structures🔗
To enable metrics computation, copy your reference CIF files to:
/apheris/input/{YOUR_BENCHMARK_NAME}/assets/{query_name}-ground-truth.cif`
Example structure:
/apheris/input/
├── cif
│ └── directory
│ ├── 5sfc.cif
│ ├── 5shh.cif
├── example-benchmark
│ └── assets
│ ├── 5sfc-ground-truth.cif
│ ├── 5shh-ground-truth.cif
Example Python snippet:
import shutil
for query_name in response.json()["queries"].keys():
shutil.copy(
f"/apheris/input/{CIF_FILE_DIRECTORY}/{query_name}.cif",
f"/apheris/input/{YOUR_BENCHMARK_NAME}/assets/{query_name}-ground-truth.cif"
)
Skip this step if you’re using your own pipeline to compute alignment metrics.
Copy all MSA files into the same /assets directory:
/apheris/input/{YOUR_BENCHMARK_NAME}/assets/
Unused MSAs in this folder will not cause issues.
Performing Prediction🔗
Run the prediction request:
response = requests.post(HUB_API_URL + "/predict", json=prediction_payload)
Your output directory will be populated with:
.heartbeat→ updated every 10 seconds during prediction.error→ present only if prediction fails.done→ created when prediction completesmeta/→ per-query JSON results with aligned CIF strings and metricsresults.zip→ raw unaligned predicted structures
Monitoring Benchmark Status🔗
Here’s a simple Python loop to monitor your job:
import time
from pathlib import Path
output_dir = Path(f"/apheris/output/{YOUR_BENCHMARK_NAME}/outputs")
while True:
if (output_dir / ".error").exists():
print(f"Benchmark {YOUR_BENCHMARK_NAME} failed.")
break
if (output_dir / "results.zip").exists():
print(f"Benchmark {YOUR_BENCHMARK_NAME} succeeded.")
break
time.sleep(10)
Note
Each prediction payload should contain only a few queries. For large benchmarks, loop over multiple smaller payloads for better performance.
Summary🔗
You’ve now learned how to:
- Query and verify OpenFold3 installations via the Hub API.
- Generate benchmark queries from CIF files.
- Prepare prediction payloads and MSAs.
- Run benchmarks and monitor outputs.
In future releases, these benchmarking workflows will be available interactively in the Apheris Hub UI.
Support and Next Steps🔗
To get access to source code, troubleshoot deployment issues, or inquire about connecting to federated environments, please contact: support@apheris.com.